Tissue microprocessing for multimodal
analysis of tumors
The objective of this project is to develop strategies to perform accurate spatial molecular profiling of heterogeneous tumor tissues and cells in culture. We aim to analyze heterogeneity at multiple spatio-temporal scales in a multi-modal approach with spatial genomic, transcriptomic and proteomic data, and evaluate the implications of the generated workflows to diagnosis, prognosis and therapeutic prediction. A particular focus has been quantification of heterogeneity from a clinical standpoint.
This work is organized as:
1. Microscale molecular assays
2. Establishing new clinical workflows
3. Enabling technologies to implement high resolution microscale molecular assays
1. Microscale molecular assays
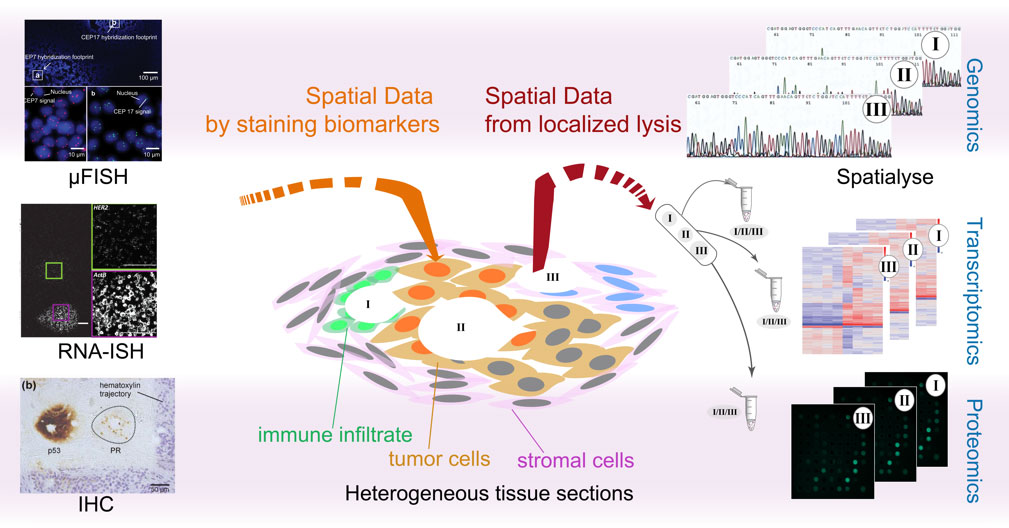
We use microfluidic technologies to implement micro-scale bio-assays, which can probe different ‘omics’, through either spatial tagging or lysis assays. We develop our assays to be inter-compatible, so as to enable a high degree of modularity and customizability. The miniaturization aspect offers the advantage of budgeting the same tissue sample for further use in a different assay (i.e., spatial multiplexing). Further, it takes advantage of the physics of fluids at the micro-scale by reducing the mass transport limitations, thus often reducing total assay times.
FISH to evaluate chromosomal aberrations
- Reduction of time and oligo probe consumption for performing fluorescence in situ
hybridization (FISH) - Development of a method to measure kinetics of FISH probe binding to validate and quantify
the efficiency of probe binding - Recirculation of the same oligo probes for reduced consumption and costs
References:
- Rapid micro fluorescence in situ hybridization in tissue sections
D. Huber, and G. V. Kaigala
Biomicrofluidics, 12(4), 042212, 2018 - Real-Time Monitoring of Fluorescence in Situ Hybridization Kinetics
N. Ostromohov, D. Hueber, M. Bercovici, G.V. Kaigala
Analytical Chemistry, 90(19), 11470-11477, 2018 - Micro fluorescence in situ hybridization (μFISH) for spatially multiplexed analysis of a cell monolayer
D. Huber, J. Autebert, and G. V. Kaigala
Biomedical Microdevices, 18(2), 40, 2016 - Fluorescence in situ hybridization (FISH): history, limitations and what to expect from micro-scale FISH?
D. Huber, L. V. von Voithenberg, G.V. Kaigala
Micro & Nano Engineering, 1, 15-24 2018
Transcriptomics
- Detection and quantification of spatial transcriptomic heterogeneity based on RNA-ISH
- Compatibility with bright-field imaging and FFPE tissue sections
- Possibility of performing bright-field spatial multiplexing on a single tissue section
References:
- Spatially multiplexed RNA in situ hybridization to reveal tumor heterogeneity
L.V. von Voithenberg, A. Fomitcheva Kharthcneko, D. Huber, G.V. Kaigala
Nucleic Acids Research, 48(3), e17, 2020
Genomics
- Highly customizable spatial lysis of tissue sections (length-scales, shear and biochemistry).
- Compatibility with a range of biological substrates, from fixed tissues, frozen tissues to cells in culture.
- Time for lysis ranges from a few seconds (DNA) to minutes (DNA and mRNA).
- High concentration and high-quality nucleic acids extracted for input into genomic (and transcriptomic) workflows
References:
- Spatially Resolved Genetic Analysis of Tissue Sections Enabled by Microscale Flow Confinement Retrieval and Isotachophoretic Purification
X.F. van Kooten, L.F.T. Petrini, A. Kashyap, L. Voith von Voithenberg, M. Bercovici, G.V. Kaigala.
Angewandte Chemie – Int Ed, 58:15259–62, 2019 - Selective local lysis and sampling of live cells for nucleic acid analysis using a microfluidic probe
A. Kashyap, J. Autebert, E. Delamarche, G.V. Kaigala
Scientific Reports 6, 29579, 2016.
Proteomics
- Increased multiplexability by analyzing neighboring microscale regions
- Reduction of staining time to <5 min, giving a time advantage of 45 min over a regular assay
- Possibility of localized dewaxing to enable archival of sections after an immunoassay has been partially performed on a section
References:
- Tissue lithography: Microscale dewaxing to enable retrospective studies on formalin-fixed paraffin- embedded (FFPE) tissue sections
J.F. Cors, A. Kashyap, A. Fomitcheva Khartchenko, P. Schraml, G.V. Kaigala.
PLoS One, 12:1–14, 2017 - Rapid micro-immunohistochemistry
R.D. Lovchik, D. Taylor, G.V. Kaigala
Microsystems Nanoengineering, 6, 2020
2. Establishing new clinical workflows
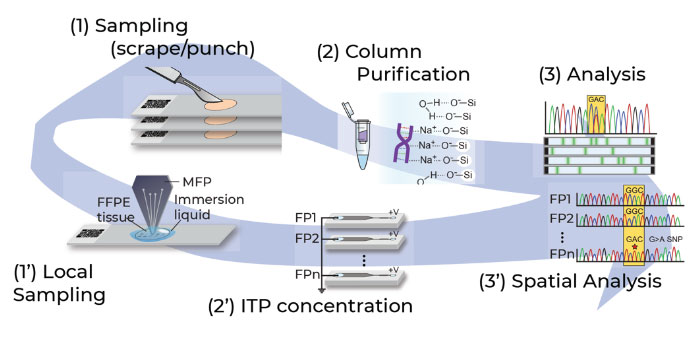
We are developing a new class of microscale molecular methods for exploring the ‘omic’ profile of cells and tissue sections, while keeping them compatible with current workflows to facilitate their integration into clinics. Such workflows are necessary to better understand the genomic and proteomic profiles of patients and potentially correlate them with patient outcome.
Spatialyse for genomics workflow
- Control over assay implementation transforms existing workflows to spatial workflows critical to profile genomic heterogeneity
- Applicable for retrospective and prospective clinical application
- Provides a spatial map of mutations in a tumor tissue section by interfacing with next generation sequencing workflows
References:
- Selective local lysis and sampling of live cells for nucleic acid analysis using a microfluidic probe
A. Kashyap, J. Autebert, E. Delamarche, G.V. Kaigala
Scientific Reports 6, 29579, 2016 - Spatially Resolved Genetic Analysis of Tissue Sections Enabled by Microscale Flow Confinement Retrieval and Isotachophoretic Purification
X.F. van Kooten, L.F.T. Petrini, A. Kashyap, L. Voith von Voithenberg, M. Bercovici, G.V. Kaigala.
Angewandte Chemie – Int Ed, 58:15259–62, 2019 - Mapping Spatial Genetic Landscapes in Tissue Sections through Microscale Integration of Sampling Methodology into Genomic Workflows
L. Voith von Voithenberg, A. Kashyap, L. Opitz, C. Aquino, T. Sykes, M. Nieser, L.F.T. Petrini, N. Enrriquez Casimiro, X.F. van Kooten, S. Biskup, R. Schlapbach, P. Schraml, G.V. Kaigala
Small, 2007901, 2021
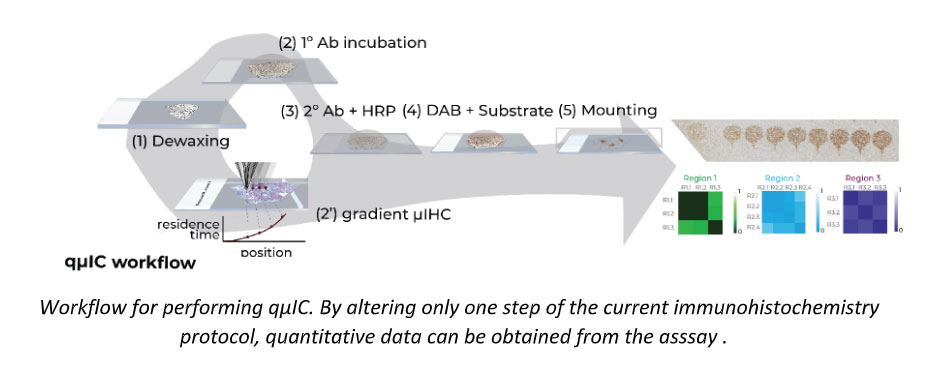
qµIC for proteomics workflow
-
Control over assay implementation transforms a non-quantitative end point assay to a spatial quantitative assay
-
Can be multiplexed for multiple proteins to obtain quantitative estimates of heterogeneity within a single tumor
-
Can be used to optimize immunohistochemical staining greatly reducing ambiguity of image-analytics based solutions
References:
- Quantitative microimmunohistochemistry for the grading of immunostains in tumor tissues,
Kashyap, A. Fomitcheva Khartchenko, P.Pati, M. Gabrani, P.Schraml and G. V. Kaigala,
Nature Biomedical Engineering, 3, 478–490, 2019.
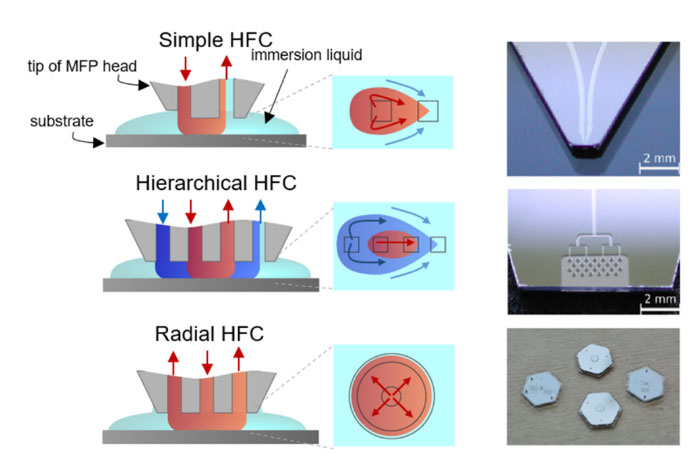
3. Enabling technologies & new mechanisms to implement microscale molecular assays
.
By using the unique physics of fluidics and chemistry at the micrometer length scales, liquids are presented to surfaces, including immersed biological surfaces. This leads to new ways of performing compartmentalization, spatio-temporal analysis, and electrokinetic concentration/separation methodologies. These are now being applied to advanced biopatterning, altering microenvironments to create unique niches, and quantifying tumor heterogeneity.
References:
- A Vertical Microfluidic Probe.
G.V. Kaigala, R.D. Lovchik, U. Drechsler, E. Delamarche
Langmuir, 2011, 27:5686–93 - Microfluidics in the “Open Space” for performing localized chemistry on biological interfaces
G.V. Kaigala, R.D. Lovchik, E. Delamarche
Angew Chemie – Int Ed, 2012, 51:11224–40 - Centimeter-scale surface interactions using hydrodynamic flow confinements
D.P. Taylor, I. Zeaf, R.D. Lovchik, G.V. Kaigala
Langmuir 32 (41), 10537-10544
